LC/MS/MS 酵母中代謝酶分析資料庫
LC/MS/MS MRM Library for Metabolic Enzymes in Yeast
包括分析代謝酵素所需的 MRM 分析條件

因應酵母菌體內大量蛋白質分析,龐大的 MRM 條件將需要被開發。
此 MRM 資料庫即包含這些分析條件,讓分析的起始準備工作變得容易。通過使用經過驗證的方法,大大減少了方法開發所需的時間。
分析酵母菌初級代謝相關酵素的理想選擇
此產品提供由 3,584 個 MRM 離子對組成的資料庫,包括穩定同位素。它涵蓋了來自出芽酵母菌的 228 種酵素,全部 498 種胰蛋白酶消化胜肽,是用於生產生質酒精或其他材料,或作為基礎研究的模式生物。此資料庫提供多種代謝途徑包括與糖解、TCA 循環、戊糖磷酸循環和胺基酸代謝的主要相關的酵素之測定。
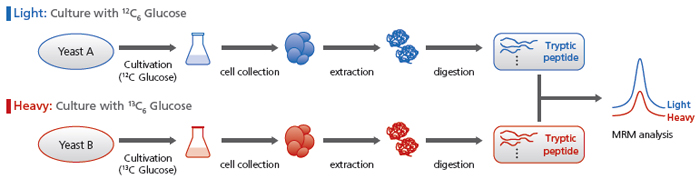
使用穩定同位素進行比較分析
列出了所有 MRM 離子對的 13C 標記胜肽的 MRM 分析條件。使用這些條件可以進行比較分析。
例如,可以將用未標記葡萄糖培養的基因缺陷酵母組與用標記葡萄糖培養的對照酵母組進行比較。
客製化方法建立
可客製化建立及選擇資料庫中特定目標分析物之分析方法。
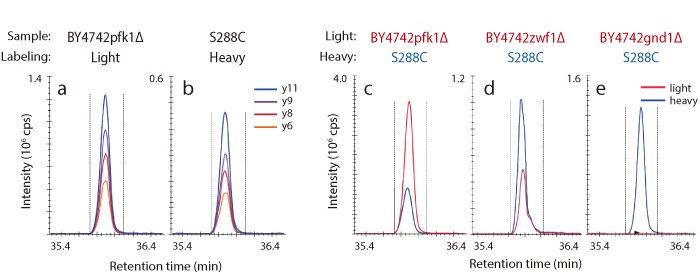
基因破壞菌株中 Gnd1p 胰蛋白酶消化胜肽的 MRM 分析
下面顯示的是用未標記的葡萄糖 (a) 生長的 BY4742pfk1Δ 菌株 (輕) 和用 13C 標記的葡萄糖 (b) 生長的 S288C 菌株 (重) 的相對層析圖。此外,基因破壞菌株中 Gnd1p 的 TIC 層析圖顯示在 (c)、(d) 和 (e) 中。在 GND1 破壞菌株中,未檢測到 Gnd1p,而在 PFK1 破壞菌株中,檢測到大量 Gnd1p。
|
參考資料: Matsuda F, Ogura T, Tomita A, Hirano I, Shimizu H. Nano-scale liquid chromatography coupled to tandem mass spectrometry using the multiple reaction monitoring mode based quantitative platform for analyzing multiple enzymes associated with central metabolic pathways ofSaccharomyces cerevisiaeusing ultra fast mass spectrometry.J Biosci Bioeng. 2015 Jan;119(1):117-20. |
代謝酵素資料庫清單
AAT1
AAT2
ACH1
ACO1
ACO2
ACS1
ACS2
ADE1
ADE12
ADE13
ADE16
ADE17
ADE2
ADE4
ADE5,7
ADE6
ADH1
ADH2
ADH3
ADH4
ADH6
ADK1
AGX1
ALD3
ALD4
ALD5
ALD6
ALT1
ALT2
ARG1
ARG2
ARG3
ARG4
ARG5,6
ARG8
ARO1
ARO2
ARO3
ARO4
ARO7
ARO8
ASN1
ASN2
ASP1
ATH1
BAT1
BAT2
BNA3
BNA5
CAR1
CAR2
CDC19
CIT1
CIT2
CIT3
CPA1
CPA2
CYS3
CYS4
DAK1
DAL7
DUR1,2
ECM17
ECM40
ENO1
ENO2
ERG10
ERG13
ERG20
ERR
FBA1
FBP1
FRD1
FUM1
GAD1
GAL1
GAL10
GAL7
GCV1
GCV2
GCY1
GDB1
GDH1
GDH2
GDH3
GLC3
GLK1
GLN1
GLT1
GLY1
GND1
GND2
GPD1
GPD2
GPH1
GPM1
GPM2
GPM3
GSY1
GSY2
GUA1
GUK1
GUT2
HIS1
HIS3
HIS4
HIS5
HIS6
HIS7
HOM2
HOM3
HOM6
HOR2
HXK1
HXK2
ICL1
IDH1
IDH2
IDP1
IDP2
IDP3
ILV1
ILV2
ILV3
ILV5
IMD2
IMD4
KGD1
KGD2
LAT1
LEU1
LEU2
LEU4
LPD1
LSC1
LSC2
LYS1
LYS12
LYS2
LYS20
LYS21
LYS4
LYS9
MAE1
MDH1
MDH2
MDH3
MET10
MET14
MET16
MET17
MET2
MET22
MET3
MET6
MHT1
MLS1
MVD1
NQM1
NTH1
PCK1
PDA1
PDB1
PDC1
PDC5
PDC6
PDE1
PFK1
PFK2
PGI1
PGK1
PGM1
PGM2
PRO1
PRO2
PRO3
PRS1
PRS2
PRS3
PRS4
PRS5
PYC1
PYC2
RHR2
RKI1
RNR2
RNR4
RPE1
SAH1
SAM1
SAM2
SAM4
SDH1
SDH2
SDH3
SDH4
SER1
SER2
SER3
SER33
SFA1
SHM2
SOL3
SOL4
TAL1
TDH1
TDH2
TDH3
THR1
THR4
TKL1
TKL2
TPI1
TPS1
TPS2
TPS3
TRP1
TRP2
TRP3
TRP5
TSL1
UGA1
UGA2
UGP1
URA2
YNK1
YPR1
ZWF1
News / Events
-
創造未來的全新飲食—「培養肉」
當你聽到「培養肉」,會聯想到什麼樣的味道呢?
或許不久的將來,我們都能親自品嚐,找出答案! -
轉發研討會資訊-食品檢驗尖端技術與應用研討會
「食品檢驗尖端技術與應用研討會」特別邀請國立陽明交通大學食品安全及健康風險評估研究所 魏國晋 副教授、中央研究院生物多樣性研究中心新世代基因體定序核心實驗室 呂美曄 博士暨研究技師、長庚大學資訊工程系暨資訊中心軟體組 李季青 副教授暨組長 (依演講順序) 齊聚一堂,為您帶來精彩的演講內容,最後由台灣島津 蔡侑達 資深應用工程師分享多維影像與精準檢測,全面解析元素至分子的食品安全。
-
2024 EDX User Meeting
我們將於 2024年9月24日於台北、2024年9月27日於高雄 舉辦電子業界實務分享會,旨在儀器教育訓練及使用者交流。
另外我們也特別邀請輻射防護回訓講師蒞臨 ,課程結束將會發放電子積分證明。 誠摯地邀請您撥冗蒞臨與指教。 -
2024食安檢驗工作坊
島津的創業宗旨為「以科學技術向社會做貢獻」,不僅是指要解決地球上各種複雜多樣的課題,也持續致力於孕育具備專業分析知識與技術的下一代。為此台灣島津將於暑假期間舉辦為期三天的化學檢驗工作坊,藉由課程及實驗操作幫助學生奠定未來學術研究及職涯發展的基礎。
-
轉發研討會資訊-2024年濫用藥物檢驗技術國際交流研討會
「濫用藥物檢驗技術國際交流研討會」由屏東縣檢驗中心及台灣島津科學儀器股份有限公司主辦,醫全實業股份有限公司協辦。特別邀請屏東縣檢驗中心 張菊香 主任、日本島津製作所 石井寿成 主任、警政署刑事警察局 謝金霖 技正、醫全實業股份有限公司 王映文 專員 (依演講順序) 齊聚一堂,為您帶來精彩的演講主題,演講內容多元精彩,誠摯邀請您撥冗蒞臨參與!
-
揭示看不見的網絡—有效分析 PFAS 的解決方案
PFAS存在於我們消費的產品、我們呼吸的空氣、我們吃的食物、我們喝的水和我們自己體內。因此,需要開發新的技術來防止它們無限期地在環境中徘徊。這些技術與 PFAS 的適當監測相結合,有助於減輕這些物質對人類生態系統的有害影響。


